Power
Problem Set 7: Parallel Sequences
Quick Links
- Code Bundle for this assignment
- Preliminaries
- Parallel Libraries
- Parallel Sequences
- Map-reduce Application
- US Census Population
- Hand-in Instructions
- Acknowledgements
Introduction
In this assignment, we will implement a functional parallel sequence abstraction on top of lower level parallel libararies for programming with futures and message passing. A key lesson is that it is possible to build high-level, reuseable parallel functional abstractions on top of lower-level imperative primitives.
This assignment has three main parts: (1) implementation of sequences, (2) implementation of a simple map-reduce app, and (3) implementation of US population queries. Parts (2) and (3) depend upon the sequence API, but we have given you a simple sequential implementation of sequences. Consequently, you do NOT have to do part (1) of the assignment first. Even if you do, we recommend that you use the simple sequential sequence implementation initially to test and debug your applications in parts (2) and (3) rather than your parallel implementation.
You may do this problem set in pairs. If you do so, both students are responsible for all components of the assignment. Please write both names at the top of all files if you do it in pairs. Please only have 1 member of the pair hand in the assignment to dropbox.
This assignment is longer and more open-ended than previous assignments. We have left you a README.txt file in which you may comment on any unusual design decisions you made or problems that you had. You are not required to write lengthy explanations of your code here but you may add any elements that you believe will help the graders understand and appreciate your work. Also, any comments on the assignment and suggestions for next year are appreciated.
Preliminaries
You will need the ocamlfind tool to compile the
handout code with the provided Makefile. Install
ocamlfind from opam with the following
command, then ensure it is in your shell's path:
opam install ocamlfind
Now, you should be able to compile your assignment using the
Makefile we included. Note that the compiler commands we have
supplied in the Makefile depend on the presence of the
_tags we have included in the tar bundle. If you delete
(or move) the _tags file, you will find that compilation
breaks, possibly with an error message that looks like this:
Error: No implementations provided for the following modules:
Unix referenced from system.cmx, future.cmx, mpi.cmx
Command exited with code 2.
Moral of the story: just don't touch the _tags file and
you should be fine. Type any of the following commands at the prompt
to build components of the assignment:
make example // Example use of the future and mpi libraries make seq // Part 1: testing your sequence implementation make seqapps // Part 2: testing the sequence applications make qpop // Part 3: testing your population queries
One difference to note from previous assignments: our Makefile
builds .native files (native machine code) rather than
.byte files (portable bytecode for the OCaml bytecode
interpreter). This switch is because in this assignment we care about
raw performance over portability.
In order to obtain the expected speedups from parallel
computation, you will need to test this assignment on a multi-core
machine. If you do not have a multi-core machine (or if you do, but
are running this on a VM that does not have access to multiple cores),
you can still implement and test all of the desired functionality. If
you have an undergraduate CS account, you can use the CS department
machines ( cycles.cs.princeton.edu ) to measure your
speedup.
(NB: we often talk about parallel "performance" but speedup is not a
perfect measure of absolute performance. The linearized version of
your algorithm (i.e., LinSeq in the test file) is just the
parallel algorithm but with SFuture instead of PFuture. A real-world
systems experiment would consider a comparison between an algorithm
designed or optimized for sequential performance versus the parallel
algorithm.)
We don't recommend using the OCaml toplevel to debug this assignment. Use the testing knowledge learned from previous assignments to write good unit test cases and infrastructure.
Parallel Libraries
In order to implement your parallel sequence abstraction, you will need to build on two lower-level parallel abstractions: a futures abstraction and a message-passing abstraction with typed channels.
Futures
A future is an abstraction that lets you run a function f in parallel and retrieve
the result later. The futures API is defined as follows:
module type FUTURE = sig
type 'a future
val force : 'a future -> 'a
val future : ('a -> 'b) -> 'a -> 'b future
end
| Function | Description |
|---|---|
future |
Start function f with parameter x in a separate process |
force |
Wait for the future computation in the separate process to return a result |
Message Passing
The message passing implementation also runs a function f
in a separate process, however it allows f to communicate
with other processes via blocking calls to send and
receive. The Mpi module defines an abstract channel type
to carry messages between processes. The type parameters
's and 'r represent the send and receive types
for the channel respectively. The API is given as follows:
module type MPI = sig
type ('s, 'r) channel
val spawn : (('r, 's) channel -> 'a -> unit) -> 'a -> ('s, 'r) channel
val send : ('s, 'r) channel -> 's -> unit
val receive : ('s, 'r) channel -> 'r
val wait_die : ('s, 'r) channel -> unit
end
| Function | Description |
|---|---|
spawn |
Run a function with its parameter in a separate process. The
function passed to spawn will be given a handle to a
channel that enables communication with the parent
process. Notice how the send and receive types for this
channel are flipped. This is because the spawned child process
will send values of the receive type 'r for the
parent and receive values of the send type 's for the
parent. The spawn function also has a return type of
unit. If you want to return a result from the child
process, you must do so via a message on the channel. |
send |
Send a value on the channel. This is a blocking send, so the
program cannot continue until someone calls receive on the
other end of the channel |
receive |
Receive a value on the channel. Likewise, this is a blocking receive. |
wait_die |
Wait for a process to finish and clean up its resources before continuing. It is VERY important that you use this in the right places. Failing to clean up dead processes, will exhaust your OS's resources and likely result in a cryptic error message. |
In the examples.ml file, you can find 2 simple
examples showing how to use the future and mpi interfaces.
Part 1: Parallel Sequences
Note: Part 1 is probably the hardest part of the assignment. You can skip part 1 at first and start with parts 2 and 3 at your discretion.
A sequence is an ordered collection of elements. OCaml's
built-in list type is one representation of sequences. In
this problem, you will develop another representation for sequences.
Your representation will be built using arrays and
the APIs for futures and message passing described above.
The following table summarizes the most important operations in the sequence module and their complexity requirements. Because we are using separate processes for parallelism, many operations like append and cons, where copying data between processes dominates any computational cost, will be faster with a purely sequential implementation. If a function is labelled as "Sequential", then you should implement it without any parallelism. The more challenging part will be to implement the functions labelled "Parallel", which can benefit from multiple processes.
The parallel functions include:
tabulate, map, reduce,
mapreduce, and scan. Ignoring copying
costs, each of these functions should have O(n) work
and O(n/p) span, where p stands for the
number of available processors. We will judge your final
implementation, in part, on the overall performance of your parallel
sequence implementation. For the purpose of performance tuning, you
may assume that we will use sequences when the amount of data is
much, much greater than the number of processors.
| Function | Description | Implementation |
|---|---|---|
tabulate |
Create a sequence of length n where each element i holds the value f(i) |
Parallel |
seq_of_array |
Create a sequence from an array | Constant time |
array_of_seq |
Create an array from a sequence | Constant time |
iter |
Iterate through the sequence applying function f on each element in order. Useful for debugging |
Sequential |
length |
Return the length of the sequence | Constant time |
empty |
Return the empty sequence | Constant time |
cons |
Return a new sequence like the old one, but with a new element at the beginning of the sequence | Sequential |
singleton |
Return the sequence with a single element | Constant time |
append |
Append two sequences together. append [a0;...;am] [b0;...;bn] = [a0;...;am;b0;...;bn]
|
Sequential |
nth |
Get the nth value in the sequence. Indexing is zero-based. |
Constant time |
map |
Map the function f over a sequence |
Parallel |
reduce |
Fold a function f over the sequence. To do this in parallel, we require that f
has type: 'a -> 'a -> 'a. Additionally, f must be associative |
Parallel |
mapreduce |
Combine the map and reduce operations. | Parallel |
flatten |
Flatten a sequence of sequences into a single sequence. flatten [[a0;a1]; [a2;a3]] = [a0;a1;a2;a3]
|
Sequential |
repeat |
Create a new sequence of length n that contains only the element provided repeat a 4 = [a;a;a;a]
|
Sequential |
zip |
Given a pair of sequences, return a sequence of pairs by drawing a value from both sequences at each shared index.
If one sequence is longer than the other, then only zip up to the last element in the shorter sequence. zip [a0;a1] [b0;b1;b2] = [(a0,b0);(a1,b1)] |
Sequential |
split |
Split a sequence at a given index and return a pair of sequences.
The value at the index should go in the second sequence. split [a0;a1;a2;a3] 1 = ([a0],[a1;a2;a3])This routine should fail if the index is beyond the limit of the sequence. |
Sequential |
scan |
This is a variation of the parallel prefix scan shown in class.
For a sequence [a0; a1; a2; ...], the result of scan will be
[f base a0; f (f base a0) a1; f (f (f base a0) a1) a2; ...] |
Parallel |
Implementation Details
One of the critical parts of the sequence implementation will be
to write parallel versions of the functions such as
tabulate, map, reduce,
mapreduce and obtain a speedup. The constant factors
will become very important in your implementation and even a
correct parallel implementation may suffer performance degradation
if you are not careful. Below are a few things to watch out for:
Array library: You may represent sequences however you would
like, but one obvious choice is to use arrays (at least in part) and
the
array library.
We've given you a suggestion for a representation in sequence.ml.
You may think our suggestion is a bit odd. It isn't the only choice and you can
change it if you would like. Notice however that it is sometimes tricky to build "empty" sequences
with alternate representations.
Recall that [||] creates an empty array. Please note
also that the sequence API must be a functional API. If your
underlying representation involves a mutable data structure such as an
array, you must make a copy of the array when implementing functions
such as map or mapreduce. You should not destructively over-write
elements of an array when performing a map.
Chunking:
In class, most of the algorithms we covered were binary
divide-and-conquer algorithms. To implement these, you must be very
careful to avoid unnecessary data copying. Consider what happens if
you try to implement a function like map over
sequences using a naïve, recursive, binary, divide-and-conquer
algorithm in conjunction with the futures library you have been
given. Your algorithm might operate by dividing the sequence in
half and copying each half in to a new process. Then it might
recursively divide each subsequence in half again, copying the
quarters in to still more processes, etc. At a certain point, the
recursion bottoms reaches a base case, and it would apply the
function across some small number of elements. On the way back up
the call stack it might copy the completed sub-parts from child
processes back in to parent processes, copying up again and again.
In other words, each array element would wind up getting copied
upwards of 2 log n times!
Instead, you should divide your sequence into some number of
chunks, and apply the map function across each chunk.
You should try to copy each element out to a new process only once
and then copy it back in only once, if you can. How many chunks
you divide the sequence into will determine a large part of the
performance of your algorithm. For example, using a single chunk
and single process will effectively run map
sequentially. Alternatively, using a chunk size of 1 will result in
an abundance of tiny computations, each of which is computed in a
separate process. In addition to running extremely slowly as the
overhead of starting up a process dominates the time to complete
its assigned computation, this will quickly exhaust the OS's
resources. You should find a happy middle ground between these two
extremes. In the sequence.ml file, there is a
num_cores value that tells you the number of cores on
your machine. We have also supplied you with a bit of code that will
divide an array up into chunks and apply a function to each chunk.
We have given an example of how to use this code in the function
tabulate.
Try running a couple of experiments to see how many
chunks works best for you. You could compare the chunking strategy we suggest
to another chunking strategy.
In your README, briefly report on your experiments and findings --- you do not need to write an opus. Include the kind of machine you used, and the number of cores available to you.
Scaling as Computational Resources Increase: Though you will
report on experiments that you did on one specific machine, your
implementation should be able to execute efficiently on machines
with different numbers of cores. So do not fix the
number of processes you use at 2 -- the number of processes used
(and performance, when the data is large) should increase as more
computational resources become available. In other words, your
algorithms should be parameterized by num_cores.
Intermediate Sequences: A naïve implementation of the
mapreduce function would first call map
and then call reduce. However, such an implementation
would create an unnecessary intermediate sequence from the map that
would subsequently be used solely for the reduce. This leads to
extra copying that can hinder performance. A better approach would
be to combine the map and reduce
functions in a single pass over the data. Be wary of creating
intermediate data structures in your implementation.
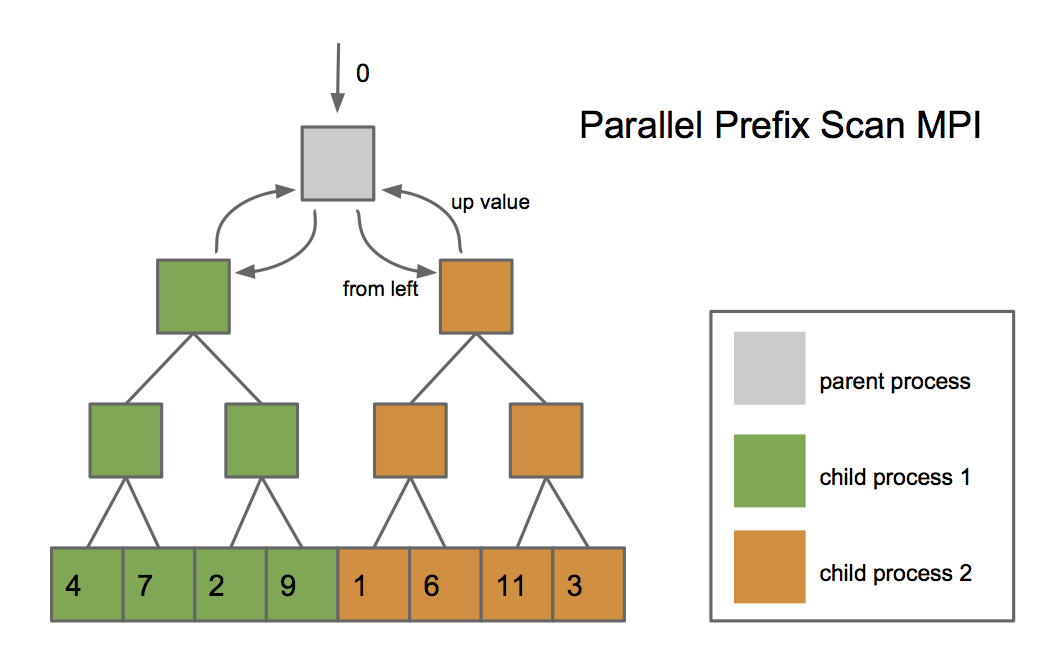
Parallel Scan Implementing the parallel prefix sum (the
scan function in sequence.ml) with good
performance is difficult without shared-memory parallelism. Recall
from class that we need to build a prefix tree in parallel in an
up phase and push the results back down in a down
phase. A naïve implementation would build the prefix tree
in the up phase by first building the subtrees in parallel
before combining them together. However, because we are using
separate processes, this would mean copying the entire subtrees
between processes. We can get around this limitation by using the
message passing interface described above. Rather than copy
subtrees between processes, we can keep the subtree in the existing
process and simply send messages between processes with the minimal
amount of information necessary for the up and
down phases of the algorithm. The image below shows what
this might look like. You must implement the
scan function using message passing. However, it may
be useful to first implement scan without message
passing and then optimize it afterwards.
Exceptions, references, and debugging, oh my! Running computations in separate processes leads to several subtle, tricky issues. For example, how should we catch and handle exceptions thrown in a different process? It is best to avoid throwing exceptions in your future computations altogether. For similar reasons, you should avoid having a mutable data structure that is written in both the parent and the child. Processes maintain the abstraction of separate address spaces, so a change to a mutable value in one process will have no effect on the same value in the main process! This means effects in your future computation will be forgotten once the future finishes.
Your Task: The details
Your task is to build an efficient implementation of sequences. We
have provided a complete interface as well as a reference
implementation based on lists in sequence.ml. This
implementation is only meant to be a guide to the functional behavior
expected of the module. The reference implementation should be useful
for debugging. You can also use this implementation to complete parts
2 and 3 of the assignment without first finishing part 1. Do be sure
to switch to your parallel sequence implementation before submitting,
though.
To test your sequence implementation, use
test_seq.ml. We have provided some infrastructure for you
to write tests for your sequence implementation. There are also a
number of benchmarks to test the performance of your sequence
implementation. The benchmarks show the speedup obtained from using
parallelism. Running the benchmarks ca. December 2014 on an 8-core
Intel i7 processor yielded these results:
Running Benchmarks, size: 1000000 ============================================== reduce : speedup=6.28587774573 map : speedup=5.90847054041 mapreduce : speedup=7.07261278358 scan : speedup=3.45738970032 ==============================================
Your results may vary depending on your computer's hardware, whether you are running in a VM, etc. Implementations with unnecessary data copying, which leads to poor performance will receive less credit.
To submit: sequence.ml
Part 2: An Inverted Index Application
The following application is designed to test out your sequence implementation and illustrate the "Map-Reduce" style of computation, as described in Google's influential paper on their distributed Map-Reduce framework.
When implementing your routines, you should do so under the assumption that your code will be executed on a parallel machine with many cores. You should also assume that you are computing over a large amount of data (e.g.: computing your inverted index may involve processing many documents; your perfect matching service may have many profiles). Hence, an implementation that iterates in sequence over all profiles or over all documents will be considered impossibly slow --- you must use bulk parallel operations such as map and reduce when processing sets of documents to minimize the span of your computation. Style and clarity is important too, particularly to aid in your own debugging.
Inverted Index
An
inverted index
is a mapping from words to the documents in which they appear. For
example, if we started with the following documents:
Document 1:
OCaml map reduce
Document 2:
fold filter ocamlThe inverted index would look like this:
| word | document |
|---|---|
| ocaml | 1 2 |
| map | 1 |
| reduce | 1 |
| fold | 2 |
| filter | 2 |
To implement this application, you should take a dataset of
documents (such as data/reuters.txt or
data/test1.txt) as input and use the map and reduce
operations to produce an inverted index. Complete the
mkindex function in
inverted_index.ml. This function should accept the
name of a data file and print the index to screen. To get started,
investigate some of the functions in the module Util
(with interface apps/util.mli and implementation
util.ml). In particular, notice that
Util contains a useful function called
load_documents --- use it to load a collection of
documents from a file. You should print the final result using
Util function print_reduce_results.
Hint: The example above suggests that your inverted index should be case insensitive (OCaml and ocaml are considered the same word). You might find functions such as String.uppercase and/or String.lowercase from the String library useful.
Your Task
Complete the mkindex functions making use
of the parallel map and reduce functionality provided by your Sequence
implementation.
To run the inverted index, first compile into an executable for testing with the given Makefile using make seqapps. Then run commands such as this one:
./main_index.native mkindex data/test1.txt
DO NOT change any code already provided to you. Make sure you place any additional functions you write in your submission files.
To submit: inverted_index.ml.
Part 3: US Census Population
The goal for this part of the assignment is to answer queries about U.S. census data from 2010. The CenPop2010.txt file in the population directory contains data for the population of roughly 220,000 geographic areas in the United States called census block groups. Each line in the file lists the latitude, longitude, and population for one such group. For the sake of simplicity, you can assume that every person counted in each group lives at the same exact latitude and longitude specified for that group.
Suppose we want to find the total population of a region in the United States. How might we go about doing this? Since we can approximate the area of any shape will sufficiently many rectangles, we will focus on simpler problem of efficiently finding the population for any rectangular area. We can think of the entire U.S. as a rectangle bounded by the minimum and maximum latitude/longitude of all the census-block-groups. The rectangle includes all of Alaska, Hawaii, Puerto Rico, and parts of Canada and the ocean. We might want to answering queries related to rectangular areas inside the U.S. such as:
- For some rectangle inside the U.S. rectangle, what is the 2010 census population total?
- For some rectangle inside the U.S. rectangle, what percentage of the total 2010 census U.S. population is in it?
We will investigate two different ways to answer queries such as these: 1) the query search method: a slower, simpler implementation that looks through every census group in parallel, and 2) the pre-computation method: a more efficient implementation that precomputes intermediate population sums to answer queries in constant time.
Query Search
We can answer a population query by searching through all the census groups and summing up the population of each census group that falls withing the query rectangle. This naïve approach must look through every single census group. However, using our parallel sequence implementation, we can improve on this by looking through census groups in parallel.
We have implemented the search-based population query in the query.ml file for you. This will serve as a reference implementation that you can use to verify your results for the precomputation-based part of the assignment that follows.
Query Precomputation
Looking at every census group each time we want to answer a query is still less than ideal. With a little extra work up front, we can answer answer queries more efficiently. We will build a data structure called a summed area table in order to answer queries in O(1) time.
Conceptually, we will overlay a grid on top of the U.S. with x columns and y rows. This is what the GUI we provide is showing you (see GUI description below). We can represent this grid of size x*y as a sequence of sequences, where each element is an int that will hold the total population for all positions that are farther South and West of that grid position. In other words, grid element g stores the total population in the rectangle whose bottom-left is the South-West corner of the country and whose upper-right corner is g.
Once we have this grid initialized, there is a neat arithmetic trick to answer queries in O(1) time. To find the total population in an area with lower-left corner (x1,y1) and upper-right corner (x2,y2), we can take the value for the top-right corner, and subtract out the values at the bottom-right and top-left corners. This leaves just the area we want, however, we have subtracted the population corresponding to the bottom-left corner twice, so we must add it back.
For this part, you must build a summed area table and answer queries by consulting the table. You should create the summed area table as follows:
- Start by initializing the grid with each element corresponding to the population just for census groups in that grid element.
- Next, build the final summed area table using appropriate operations from the parallel sequence library.
You will implement the precompute and population_lookup functions
in query.ml. You can now test your results by following the instructions
in the Testing and Visualization section below. Your results from the search-based
population query and the precomputation-based population query should match.
Testing and Visualization
We have provided the parse.ml and population.ml files
to read the census data and answer queries via command line arguments passed to the program.
The format for a query is:
./population.native [file] [rows] [cols] [left] [bottom] [right] [top]
The number of rows and columns are used for summed area table. Left, bottom, right, and top describe the row and column grid indices (starting at 1) that define the rectangular query. For example, to query a the section of the central U.S. shown highlighted in the picture below using a 20 row 40 column grid, you would run the following command:
./population.native population/CenPop2010.txt 20 40 26 4 30 7 40821662,13.1 40821662,13.1
The program will evaluate the query using both the implementations described above and list the total population and percent of the U.S. population for both. To make testing easier, we provide a GUI that displays a map of the U.S. that you can interact with. You can change the number of rows and columns, select a region on the map, and ask the GUI to run your program to display the answer.
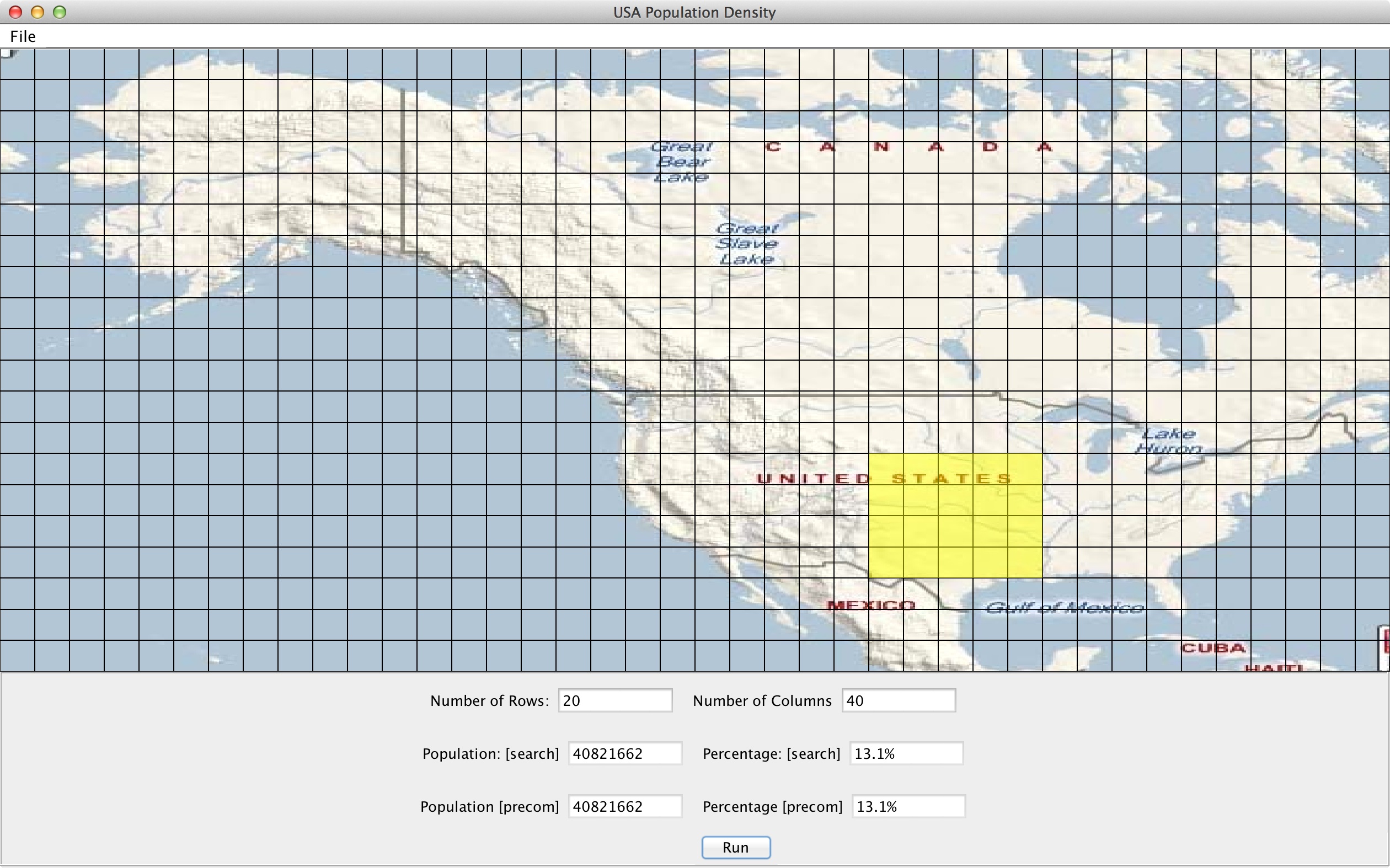
Start the GUI by running:
java -jar USMap.jar
To help you debug your code, there are a number of example queries and results listed below. Your answer should roughly match up with the search-based implementation. It is ok if you get slightly different answers (depending on how you handle edge cases), but make sure that you are at least accurate to the nearest percent.
# Population of the western half of the United States ./population.native population/CenPop2010.txt 10 10 1 1 6 10 64620478,20.7 64620478,20.7 # Population of the mainland United States ./population.native population/CenPop2010.txt 100 200 89 8 191 45 305896552,97.9 305896552,97.9 # Population of Alaska ./population.native population/CenPop2010.txt 2 1 1 2 1 2 710231,0.2 710231,0.2
To submit: query.ml
Handin Instructions
You must hand in these files to dropbox (see link on assignment page):
-
README.txt -
sequence.ml -
inverted_index.ml -
query.ml
Acknowledgments
Parts 1 and 2 of this assignment are based on materials developed by Dan Licata and David Bindel originally, and modified by Nate Foster. Part 3 is based on material developed by Dan Grossman. Ryan Beckett synthesized ideas from these sources and created the current version of the assignment.